MacroAPE 1083:6-AGGAAGTGAYGYAA
From FANTOM5_SSTAR
Full Name: 6-AGGAAGTGAYGYAA,CNhs13488,BestGuess:PU.1(ETS)/ThioMac-PU.1-ChIP-Seq/Homer(0.743657150708996),T:2870.0(34.51%),B:9265.3(22.80%),P:1e-129
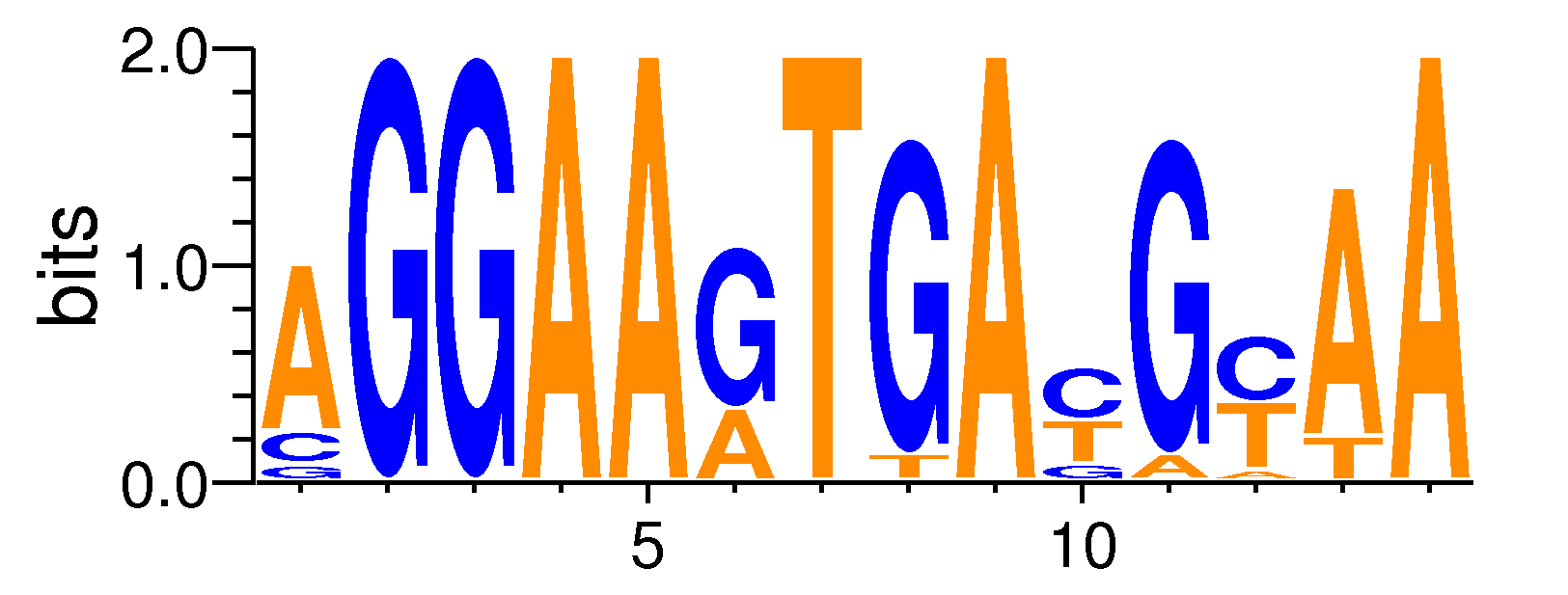
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| SPI1#MA0080.2 | 0 | 1.47881e-05 | 0.00703912 | 0.0140263 | 7 | AGGAAGTGACGTAA | AGGAAGT | + |
| FEV#MA0156.1 | 1 | 0.000306631 | 0.145956 | 0.145417 | 7 | AGGAAGTGACGTAA | CAGGAAAT | + |
| ELF5#MA0136.1 | 1 | 0.000631339 | 0.300517 | 0.183289 | 8 | AGGAAGTGACGTAA | AAGGAAGTA | - |
| GABPA#MA0062.2 | 1 | 0.000772978 | 0.367937 | 0.183289 | 10 | AGGAAGTGACGTAA | CCGGAAGTGGC | + |
| ETS1#MA0098.1 | 0 | 0.0018957 | 0.902352 | 0.345354 | 6 | AGGAAGTGACGTAA | CGGAAA | - |
| SPI1#MA0080.2 | 0 | 0.00218467 | 1.0399 | 0.345354 | 6 | AGGAAGTGACGTAA | CGGAAG | + |
| CST6#MA0286.1 | -5 | 0.00313925 | 1.49428 | 0.425361 | 9 | AGGAAGTGACGTAA | ATGACGTAA | + |
| GCR2#MA0305.1 | 0 | 0.0056034 | 2.66722 | 0.664342 | 7 | AGGAAGTGACGTAA | AGGAAGC | - |
| ABF1#MA0265.1 | 4 | 0.0069566 | 3.31134 | 0.667637 | 12 | AGGAAGTGACGTAA | TCGTATAAAGTGATAA | + |
| Eip74EF#MA0026.1 | 1 | 0.00748608 | 3.56337 | 0.667637 | 6 | AGGAAGTGACGTAA | CCGGAAG | + |
| GABPA#MA0062.2 | 2 | 0.00774288 | 3.68561 | 0.667637 | 8 | AGGAAGTGACGTAA | ACCGGAAGAG | + |
| TGA1A#MA0129.1 | -6 | 0.00885511 | 4.21503 | 0.677451 | 7 | AGGAAGTGACGTAA | TGACGTA | - |
| CREB1#MA0018.2 | -6 | 0.00928519 | 4.41975 | 0.677451 | 8 | AGGAAGTGACGTAA | TGACGTCA | + |
| YAP1#MA0415.1 | 0 | 0.0117538 | 5.59483 | 0.79631 | 14 | AGGAAGTGACGTAA | ACGAGCTTACGTAAGCAAAT | - |
| CREB1#MA0018.2 | -1 | 0.0151546 | 7.21357 | 0.810348 | 12 | AGGAAGTGACGTAA | CCTGGTGACGTC | + |
| ELK4#MA0076.1 | 2 | 0.0160664 | 7.6476 | 0.810348 | 7 | AGGAAGTGACGTAA | ACCGGAAGT | + |
| SPIB#MA0081.1 | 2 | 0.0161392 | 7.68224 | 0.810348 | 5 | AGGAAGTGACGTAA | AGAGGAA | + |
| NFIL3#MA0025.1 | -3 | 0.0163344 | 7.7752 | 0.810348 | 11 | AGGAAGTGACGTAA | ATGTTACATAA | - |
| gt#MA0447.1 | -5 | 0.0177489 | 8.44846 | 0.810348 | 9 | AGGAAGTGACGTAA | ATTACGTAAT | + |
| CBF1#MA0281.1 | -1 | 0.0187443 | 8.92227 | 0.810348 | 8 | AGGAAGTGACGTAA | GCACGTGA | + |
| NR3C1#MA0113.1 | 2 | 0.0196503 | 9.35354 | 0.810348 | 14 | AGGAAGTGACGTAA | TCAGGACATAATGTTCCC | - |