MacroAPE 1083:PLAG1 si
From FANTOM5_SSTAR
Full Name: PLAG1_si
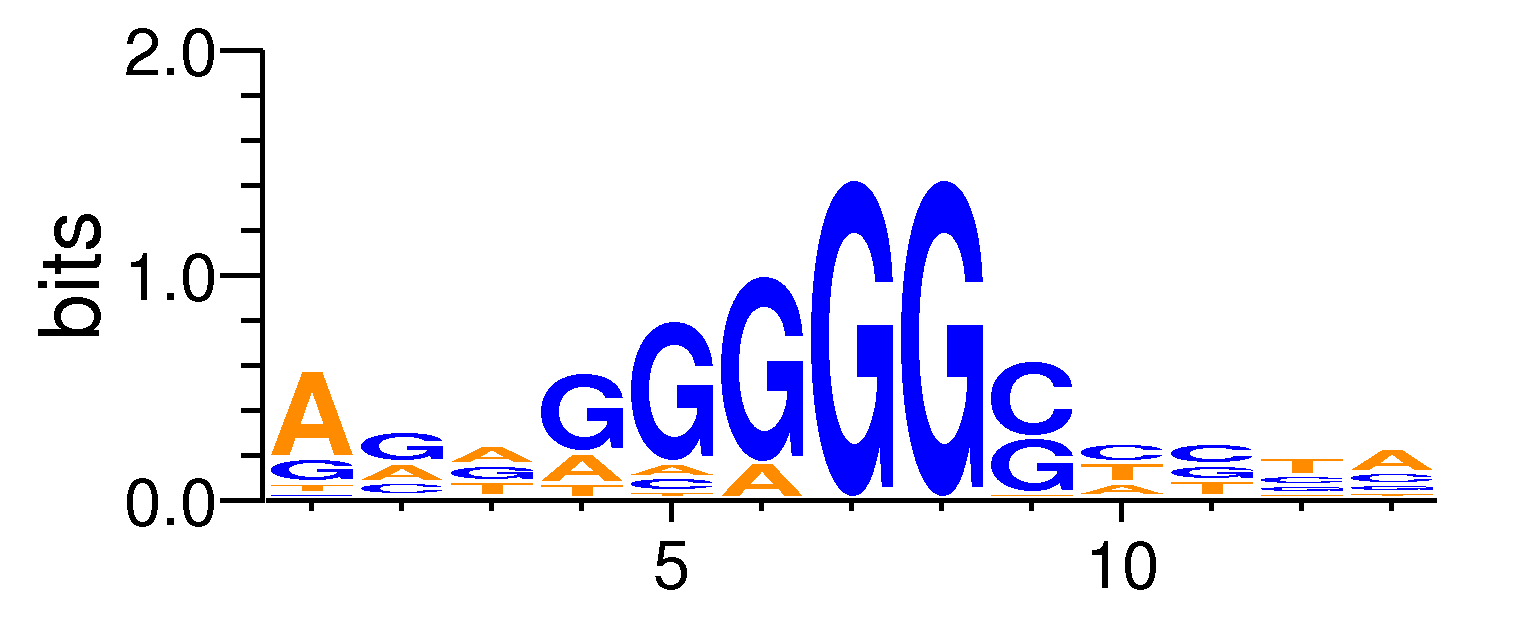
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Macho-1#MA0118.1 | -2 | 0.000256114 | 0.12191 | 0.182071 | 9 | AGAGGGGGCCCTA | TGGGGGGTC | + |
| MSN2#MA0341.1 | -3 | 0.000411363 | 0.195809 | 0.182071 | 5 | AGAGGGGGCCCTA | AGGGG | + |
| opa#MA0456.1 | 1 | 0.000579405 | 0.275797 | 0.182071 | 11 | AGAGGGGGCCCTA | CAGCGGGGGGTC | - |
| Pax4#MA0068.1 | 1 | 0.00101564 | 0.483446 | 0.239365 | 13 | AGAGGGGGCCCTA | GGGGGGGGAGTGGAGTATTGGAAATTTTTC | - |
| MSN4#MA0342.1 | -3 | 0.00153481 | 0.730567 | 0.289376 | 5 | AGAGGGGGCCCTA | AGGGG | + |
| MIG1#MA0337.1 | -1 | 0.00264584 | 1.25942 | 0.303897 | 7 | AGAGGGGGCCCTA | GCGGGGG | - |
| YML081W#MA0431.1 | -1 | 0.00271133 | 1.2906 | 0.303897 | 9 | AGAGGGGGCCCTA | GTGCGGGGT | - |
| PLAG1#MA0163.1 | -4 | 0.00312798 | 1.48892 | 0.303897 | 9 | AGAGGGGGCCCTA | GGGGCCCAAGGGGG | + |
| RGM1#MA0366.1 | -3 | 0.00377987 | 1.79922 | 0.303897 | 5 | AGAGGGGGCCCTA | AGGGG | + |
| YGR067C#MA0425.1 | 4 | 0.00393051 | 1.87092 | 0.303897 | 10 | AGAGGGGGCCCTA | TGAAAAAGTGGGGT | - |
| YER130C#MA0423.1 | 0 | 0.00394259 | 1.87667 | 0.303897 | 9 | AGAGGGGGCCCTA | AATAGGGGT | - |
| CTCF#MA0139.1 | 6 | 0.0040969 | 1.95013 | 0.303897 | 13 | AGAGGGGGCCCTA | TGGCCACCAGGGGGCGCTA | + |
| CRZ1#MA0285.1 | -4 | 0.00419073 | 1.99479 | 0.303897 | 9 | AGAGGGGGCCCTA | GTGGCTTAG | - |
| ADR1#MA0268.1 | -1 | 0.00452801 | 2.15533 | 0.304901 | 7 | AGAGGGGGCCCTA | GTGGGGT | - |
| btd#MA0443.1 | -3 | 0.00538822 | 2.56479 | 0.338636 | 10 | AGAGGGGGCCCTA | AGGGGGCGGA | + |
| SP1#MA0079.2 | 2 | 0.00640972 | 3.05103 | 0.377658 | 8 | AGAGGGGGCCCTA | GGGGGCGGGG | - |
| MIG2#MA0338.1 | -2 | 0.0080092 | 3.81238 | 0.400737 | 7 | AGAGGGGGCCCTA | TGCGGGG | - |
| SP1#MA0079.2 | 0 | 0.0080402 | 3.82713 | 0.400737 | 10 | AGAGGGGGCCCTA | GGGGCGGGGT | + |
| GIS1#MA0306.1 | 0 | 0.00847374 | 4.0335 | 0.400737 | 9 | AGAGGGGGCCCTA | TTTAGGGGT | - |
| abi4#MA0123.1 | -3 | 0.00850178 | 4.04685 | 0.400737 | 10 | AGAGGGGGCCCTA | GGGGGCACCG | - |
| AFT1#MA0269.1 | 3 | 0.0112566 | 5.35815 | 0.505322 | 13 | AGAGGGGGCCCTA | CCAATCGGGTGCAATATGTAA | - |
| MZF1_1-4#MA0056.1 | -2 | 0.0127273 | 6.05818 | 0.545371 | 6 | AGAGGGGGCCCTA | TGGGGA | + |
| STP2#MA0395.1 | 1 | 0.014006 | 6.66687 | 0.570393 | 13 | AGAGGGGGCCCTA | TGATCGGCGCCGCACGACGA | + |
| Klf4#MA0039.1 | 2 | 0.0153725 | 7.31733 | 0.570393 | 8 | AGAGGGGGCCCTA | TAAAGGAAGG | + |
| YPR022C#MA0436.1 | -1 | 0.0154259 | 7.34273 | 0.570393 | 7 | AGAGGGGGCCCTA | CGTGGGG | - |
| MZF1_5-13#MA0057.1 | -1 | 0.0161912 | 7.70703 | 0.570393 | 10 | AGAGGGGGCCCTA | GGAGGGGGAA | + |
| MIG3#MA0339.1 | -2 | 0.0163365 | 7.77616 | 0.570393 | 7 | AGAGGGGGCCCTA | TGCGGGG | - |