MacroAPE 1083:CNhs10636 bu f0
From FANTOM5_SSTAR
Full Name: CNhs10636_bu_f0
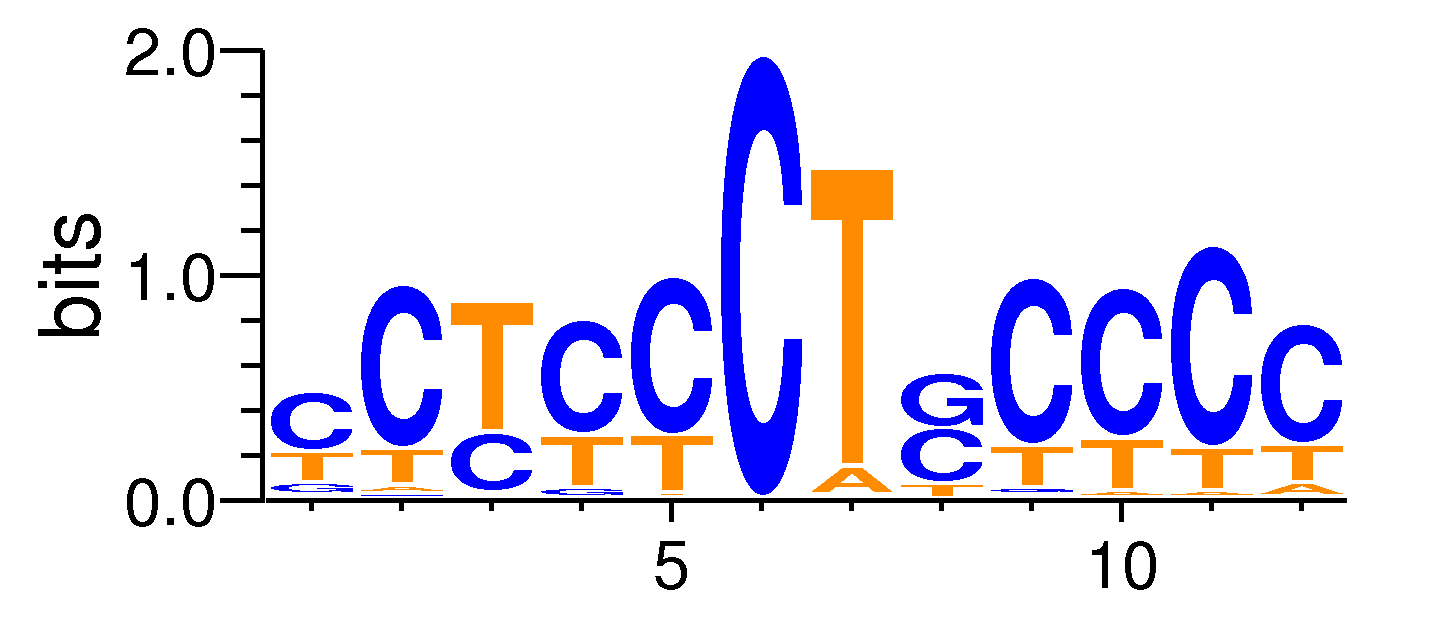
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| SP1#MA0079.2 | -2 | 5.04218e-05 | 0.0240008 | 0.0478881 | 10 | CCTCCCTGCCCC | CCCCGCCCCC | + |
| SP1#MA0079.2 | -2 | 0.000494523 | 0.235393 | 0.234836 | 10 | CCTCCCTGCCCC | ACCCCGCCCC | - |
| Pax4#MA0068.1 | 17 | 0.00206328 | 0.982119 | 0.497858 | 12 | CCTCCCTGCCCC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| RXR::RAR_DR5#MA0159.1 | 5 | 0.00252204 | 1.20049 | 0.497858 | 12 | CCTCCCTGCCCC | TGACCTCTCCGTGACCT | - |
| ESR1#MA0112.2 | 8 | 0.002621 | 1.2476 | 0.497858 | 12 | CCTCCCTGCCCC | GGCCCAGGTCACCCTGACCT | + |
| btd#MA0443.1 | -3 | 0.0035054 | 1.66857 | 0.529408 | 9 | CCTCCCTGCCCC | TCCGCCCCCT | - |
| PPARG::RXRA#MA0065.2 | 1 | 0.00390194 | 1.85732 | 0.529408 | 12 | CCTCCCTGCCCC | TGACCTTTGCCCTAC | - |
| Trl#MA0205.1 | 0 | 0.00666285 | 3.17152 | 0.779594 | 10 | CCTCCCTGCCCC | TTGCTCTCTC | + |
| Klf4#MA0039.1 | -3 | 0.00738758 | 3.51649 | 0.779594 | 9 | CCTCCCTGCCCC | CCTTCCTTTA | - |
| Tal1::Gata1#MA0140.1 | 4 | 0.00899819 | 4.28314 | 0.796776 | 12 | CCTCCCTGCCCC | CTTATCTGTCCCCACCAG | - |
| Pax5#MA0014.1 | 8 | 0.00922827 | 4.39266 | 0.796776 | 12 | CCTCCCTGCCCC | CCGCTACGCTTCAGTGCTCT | - |
| EWSR1-FLI1#MA0149.1 | 0 | 0.0108468 | 5.16307 | 0.858477 | 12 | CCTCCCTGCCCC | CCTTCCTTCCTTCCTTCC | - |
| RGM1#MA0366.1 | -2 | 0.0124857 | 5.9432 | 0.911077 | 5 | CCTCCCTGCCCC | CCCCT | - |
| MZF1_5-13#MA0057.1 | 1 | 0.0141058 | 6.71438 | 0.911077 | 9 | CCTCCCTGCCCC | TTCCCCCTAC | - |
| MZF1_1-4#MA0056.1 | -6 | 0.0160104 | 7.62095 | 0.911077 | 6 | CCTCCCTGCCCC | TCCCCA | - |
| ELK1#MA0028.1 | -1 | 0.0170687 | 8.12469 | 0.911077 | 10 | CCTCCCTGCCCC | CTTCCGGCTC | - |
| abi4#MA0123.1 | -3 | 0.0170687 | 8.12469 | 0.911077 | 9 | CCTCCCTGCCCC | CGGTGCCCCC | + |
| GAL4#MA0299.1 | 2 | 0.0172671 | 8.21913 | 0.911077 | 12 | CCTCCCTGCCCC | GAGTACTGTTCCCCG | - |
| CHA4#MA0283.1 | -4 | 0.0200373 | 9.53777 | 0.98721 | 8 | CCTCCCTGCCCC | TCTCCGCC | - |