MacroAPE 1083:CNhs12952 bu f0
From FANTOM5_SSTAR
Full Name: CNhs12952_bu_f0
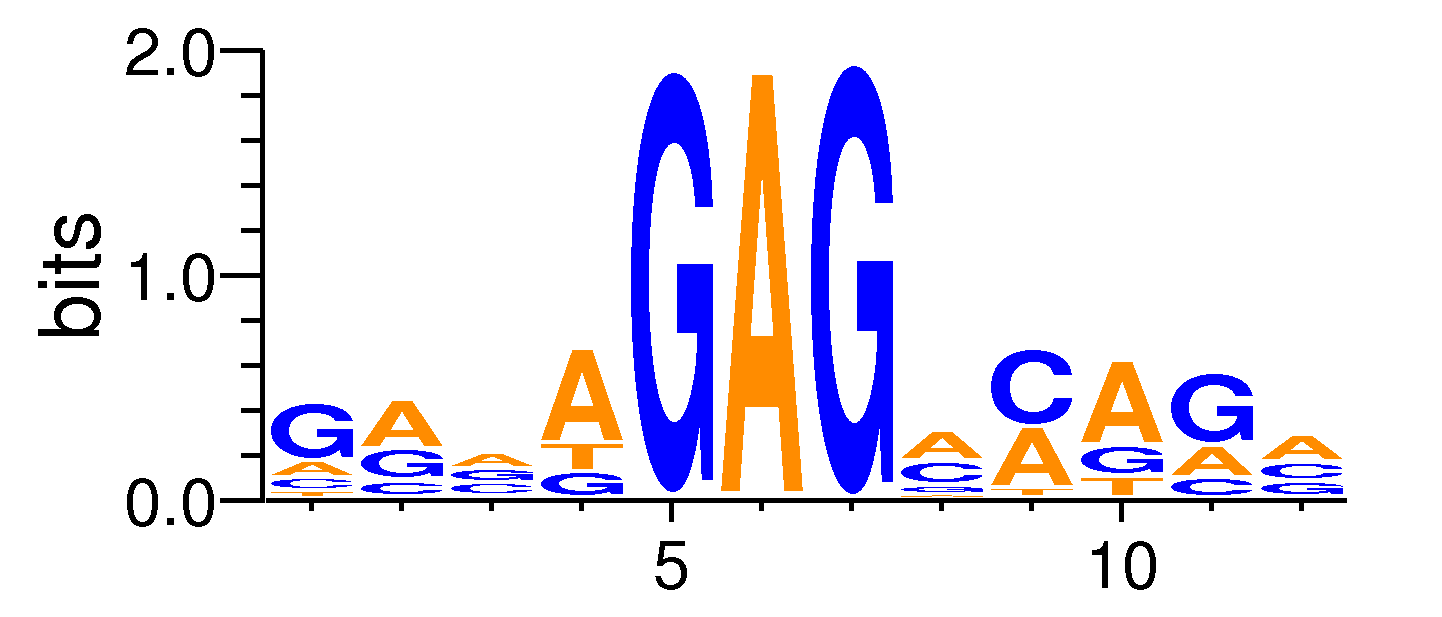
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Trl#MA0205.1 | 0 | 1.05189e-06 | 0.000500702 | 0.0010014 | 10 | GAAAGAGACAGA | GAGAGAGCAA | - |
| id1#MA0120.1 | 1 | 0.00148042 | 0.70468 | 0.70468 | 11 | GAAAGAGACAGA | CGAAAAGGAAAA | - |
| Gata1#MA0035.2 | -1 | 0.00384051 | 1.82808 | 1 | 11 | GAAAGAGACAGA | ACAGATAAGAA | + |
| BAS1#MA0278.1 | 5 | 0.0102192 | 4.86433 | 1 | 12 | GAAAGAGACAGA | GCACAGCCAGAGTCAAGTCAA | + |
| ECM23#MA0293.1 | -1 | 0.0106395 | 5.06441 | 1 | 11 | GAAAGAGACAGA | AAAGATCTAAA | + |
| GAT4#MA0302.1 | -1 | 0.011256 | 5.35787 | 1 | 11 | GAAAGAGACAGA | AAAGATCTAAA | + |
| Tal1::Gata1#MA0140.1 | 2 | 0.0148595 | 7.07314 | 1 | 12 | GAAAGAGACAGA | CTGGTGGGGACAGATAAG | + |
| IRF1#MA0050.1 | 0 | 0.0160878 | 7.65779 | 1 | 12 | GAAAGAGACAGA | GAAAGCGAAACC | + |
| Klf4#MA0039.1 | -2 | 0.0198954 | 9.47021 | 1 | 10 | GAAAGAGACAGA | TAAAGGAAGG | + |