MacroAPE 1083:ZEP2 si
From FANTOM5_SSTAR
Full Name: ZEP2_si
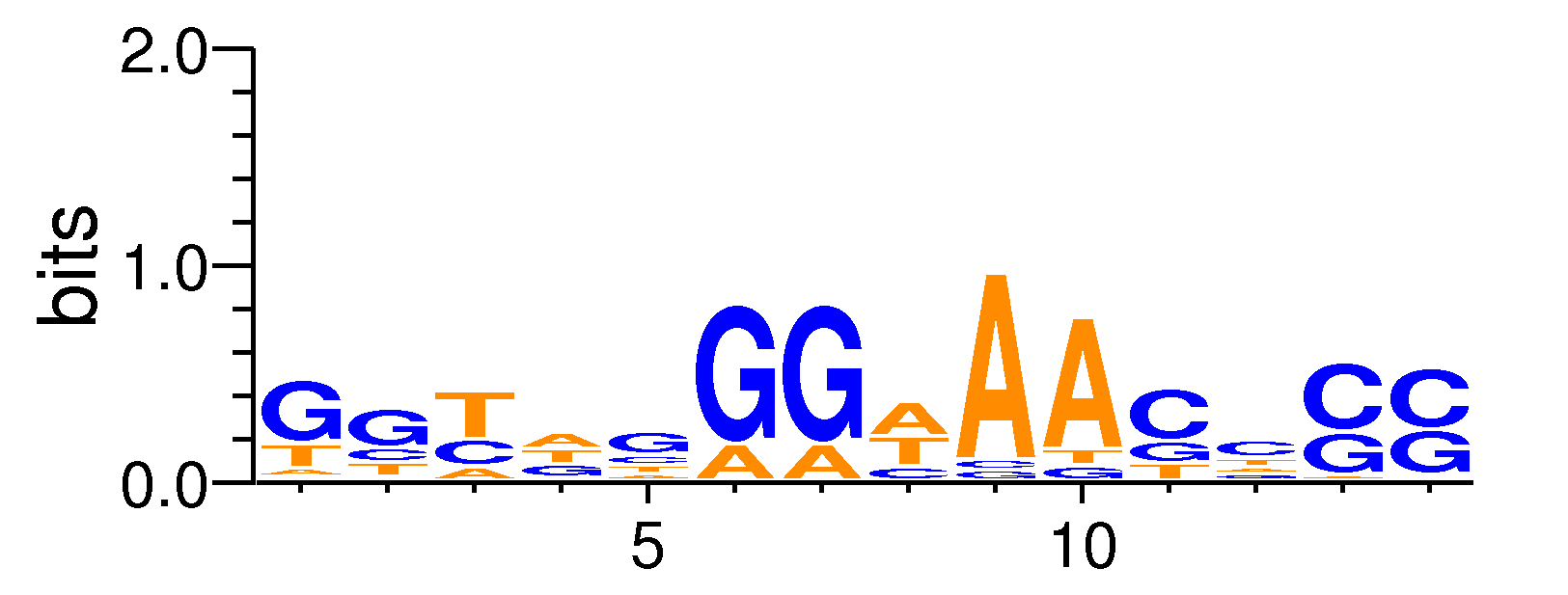
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| REL#MA0101.1 | -5 | 0.000286499 | 0.136373 | 0.27055 | 9 | GGTAGGGAAACCCC | GGAAATCCCC | - |
| RELA#MA0107.1 | -5 | 0.0014015 | 0.667116 | 0.517459 | 9 | GGTAGGGAAACCCC | GGAAATTCCC | - |
| Spz1#MA0111.1 | -3 | 0.00234274 | 1.11514 | 0.517459 | 11 | GGTAGGGAAACCCC | AGGGTAACAGC | + |
| dl_2#MA0023.1 | -4 | 0.00291634 | 1.38818 | 0.517459 | 10 | GGTAGGGAAACCCC | GGAAAACCCC | - |
| REB1#MA0363.1 | -2 | 0.00303496 | 1.44464 | 0.517459 | 9 | GGTAGGGAAACCCC | CCGGGTAAC | - |
| NFKB1#MA0105.1 | -4 | 0.00380379 | 1.8106 | 0.517459 | 10 | GGTAGGGAAACCCC | GGGGAATCCCC | - |
| GSM1#MA0308.1 | -1 | 0.00414855 | 1.97471 | 0.517459 | 13 | GGTAGGGAAACCCC | ATTAAAAAACTCCGGAGTATA | + |
| RFX1#MA0365.1 | -4 | 0.00488987 | 2.32758 | 0.517459 | 8 | GGTAGGGAAACCCC | TGGCAACC | - |
| MZF1_5-13#MA0057.1 | 0 | 0.00541105 | 2.57566 | 0.517459 | 10 | GGTAGGGAAACCCC | GGAGGGGGAA | + |
| ZMS1#MA0441.1 | 0 | 0.00547964 | 2.60831 | 0.517459 | 9 | GGTAGGGAAACCCC | TGCGGGGAA | - |
| NF-kappaB#MA0061.1 | -4 | 0.00646164 | 3.07574 | 0.55472 | 10 | GGTAGGGAAACCCC | GGAAATTCCC | - |
| HLF#MA0043.1 | 0 | 0.00710438 | 3.38168 | 0.559073 | 12 | GGTAGGGAAACCCC | TATTACGTAACC | - |
| dl_1#MA0022.1 | -2 | 0.00790183 | 3.76127 | 0.573995 | 12 | GGTAGGGAAACCCC | CGGAAAAACCCC | - |
| znf143#MA0088.1 | 8 | 0.0101139 | 4.81422 | 0.645825 | 12 | GGTAGGGAAACCCC | GCAAGGCATGATGGGAAATC | - |
| Cebpa#MA0102.1 | -2 | 0.0102585 | 4.88303 | 0.645825 | 12 | GGTAGGGAAACCCC | TTTCGCAATCTT | + |
| che-1#MA0260.1 | -6 | 0.0122097 | 5.8118 | 0.71333 | 6 | GGTAGGGAAACCCC | GAAACC | + |
| Gamyb#MA0034.1 | -5 | 0.0134141 | 6.38512 | 0.71333 | 9 | GGTAGGGAAACCCC | GACAACCGCC | + |
| ESR1#MA0112.2 | 1 | 0.0135969 | 6.47211 | 0.71333 | 14 | GGTAGGGAAACCCC | GGCCCAGGTCACCCTGACCT | + |
| Pax4#MA0068.1 | 8 | 0.014525 | 6.91389 | 0.721914 | 14 | GGTAGGGAAACCCC | GGGGGGGGAGTGGAGTATTGGAAATTTTTC | - |
| Ar#MA0007.1 | 0 | 0.0180023 | 8.5691 | 0.762943 | 14 | GGTAGGGAAACCCC | GGCGGGTACACGATGTTCTTAT | - |