MacroAPE 1083:ZN350 f1
From FANTOM5_SSTAR
Full Name: ZN350_f1
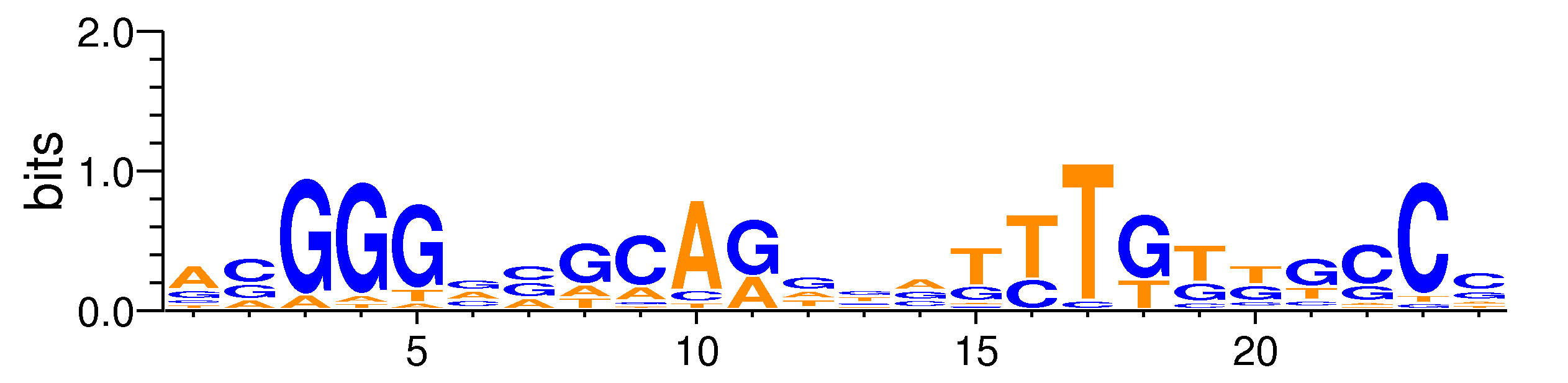
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Pax4#MA0068.1 | 0 | 0.000100961 | 0.0480576 | 0.0718816 | 24 | ACGGGGCGCAGGGATTTGTTGCCC | GGGGGGGGAGTGGAGTATTGGAAATTTTTC | - |
| GAL4#MA0299.1 | -1 | 0.00015137 | 0.072052 | 0.0718816 | 15 | ACGGGGCGCAGGGATTTGTTGCCC | CGGGGAACAGTACTC | + |
| MSN2#MA0341.1 | 0 | 0.00234088 | 1.11426 | 0.538108 | 5 | ACGGGGCGCAGGGATTTGTTGCCC | AGGGG | + |
| Macho-1#MA0118.1 | -1 | 0.00270291 | 1.28659 | 0.538108 | 9 | ACGGGGCGCAGGGATTTGTTGCCC | TGGGGGGTC | + |
| MSN4#MA0342.1 | 0 | 0.00283289 | 1.34846 | 0.538108 | 5 | ACGGGGCGCAGGGATTTGTTGCCC | AGGGG | + |
| RGM1#MA0366.1 | 0 | 0.00409333 | 1.94843 | 0.640553 | 5 | ACGGGGCGCAGGGATTTGTTGCCC | AGGGG | + |
| RREB1#MA0073.1 | -2 | 0.00472111 | 2.24725 | 0.640553 | 20 | ACGGGGCGCAGGGATTTGTTGCCC | TGGGGGGGGGTGGTTTGGGG | - |
| Pax5#MA0014.1 | -4 | 0.00610923 | 2.908 | 0.72528 | 20 | ACGGGGCGCAGGGATTTGTTGCCC | GGAGCACTGAAGCGTAACCG | + |
| btd#MA0443.1 | 0 | 0.00757885 | 3.60753 | 0.771028 | 10 | ACGGGGCGCAGGGATTTGTTGCCC | AGGGGGCGGA | + |
| NDT80#MA0343.1 | -7 | 0.00811823 | 3.86428 | 0.771028 | 17 | ACGGGGCGCAGGGATTTGTTGCCC | TTGCGTTTTTGTGGCCGGAAA | - |
| abi4#MA0123.1 | -1 | 0.00905203 | 4.30877 | 0.78156 | 10 | ACGGGGCGCAGGGATTTGTTGCCC | GGGGGCACCG | - |
| En1#MA0027.1 | -12 | 0.0125388 | 5.96849 | 0.916058 | 11 | ACGGGGCGCAGGGATTTGTTGCCC | AAGTAGTGCCC | + |
| SFP1#MA0378.1 | -3 | 0.0151453 | 7.20915 | 0.997635 | 21 | ACGGGGCGCAGGGATTTGTTGCCC | ATTGAAAAAAATTTTCTACGG | + |
| Gfi#MA0038.1 | -8 | 0.0199915 | 9.51593 | 0.997635 | 10 | ACGGGGCGCAGGGATTTGTTGCCC | CAGTGATTTG | - |
| GCR2#MA0305.1 | -2 | 0.0208118 | 9.90641 | 0.997635 | 7 | ACGGGGCGCAGGGATTTGTTGCCC | AGGAAGC | - |