MacroAPE 1083:6-ATATTTGGCA
From FANTOM5_SSTAR
Full Name: 6-ATATTTGGCA,BestGuess:MA0161.1_NFIC(0.742456156469808),T:660.0(16.57%),B:4812.3(10.56%),P:1e-27
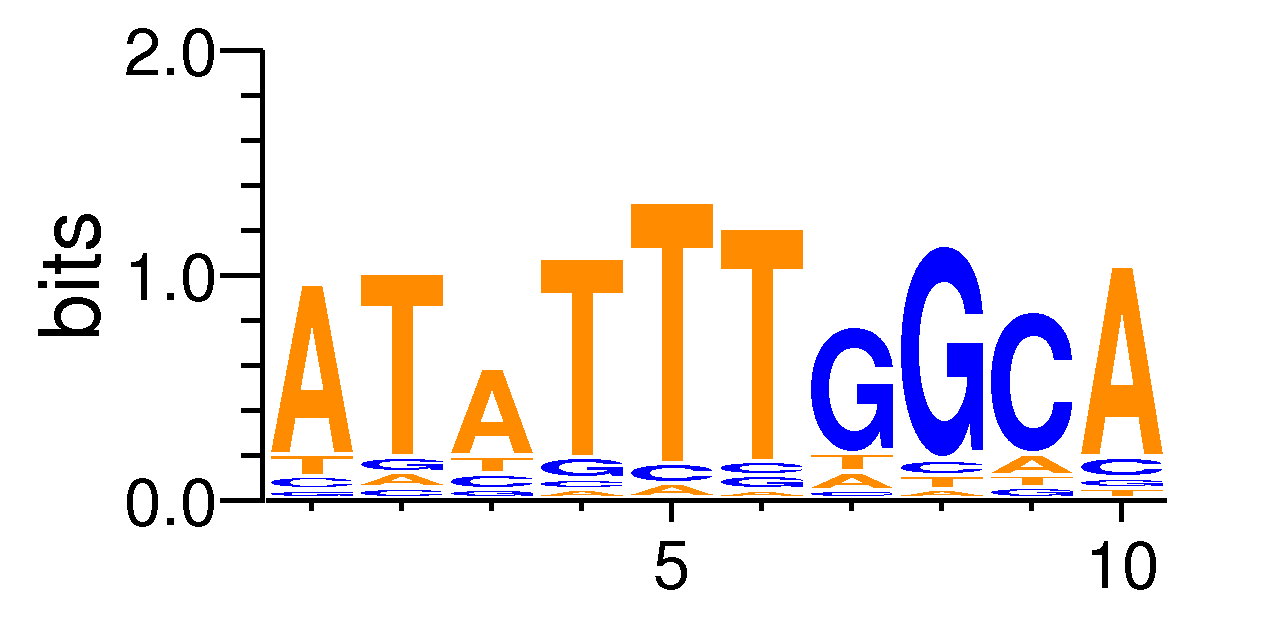
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| NFIC#MA0161.1 | -4 | 0.000153136 | 0.0728928 | 0.145786 | 6 | ATATTTGGCA | TTGGCA | + |
| AGL3#MA0001.1 | 2 | 0.0022661 | 1.07866 | 0.763749 | 8 | ATATTTGGCA | CTATTTATGG | - |
| HAP2#MA0313.1 | -4 | 0.00240677 | 1.14562 | 0.763749 | 5 | ATATTTGGCA | TTGGT | + |
| exd#MA0222.1 | -2 | 0.00563075 | 2.68024 | 0.902583 | 8 | ATATTTGGCA | TTTTGACA | + |
| ATHB-5#MA0110.1 | 1 | 0.00569321 | 2.70997 | 0.902583 | 8 | ATATTTGGCA | AATAATTGG | - |
| Nobox#MA0125.1 | -1 | 0.0090587 | 4.31194 | 0.902583 | 8 | ATATTTGGCA | TAATTGGT | + |
| Dr#MA0188.1 | -1 | 0.00927011 | 4.41257 | 0.902583 | 7 | ATATTTGGCA | TAATTGG | - |
| squamosa#MA0082.1 | 6 | 0.00973561 | 4.63415 | 0.902583 | 8 | ATATTTGGCA | ATTTCCATTTTTGG | - |
| RIM101#MA0368.1 | -3 | 0.0101189 | 4.81659 | 0.902583 | 7 | ATATTTGGCA | CTTGGCG | - |
| nub#MA0453.1 | 0 | 0.0114468 | 5.44869 | 0.902583 | 10 | ATATTTGGCA | CTAATTTGCATA | - |
| AG#MA0005.1 | 2 | 0.0115423 | 5.49416 | 0.902583 | 9 | ATATTTGGCA | CCAAATTTGGT | + |
| Hand1::Tcfe2a#MA0092.1 | -1 | 0.0123853 | 5.89542 | 0.902583 | 9 | ATATTTGGCA | GGTCTGGCAT | + |
| TOS8#MA0408.1 | -3 | 0.0154074 | 7.33392 | 0.977855 | 7 | ATATTTGGCA | TTTGACAG | - |
| E2F1#MA0024.1 | -3 | 0.0172106 | 8.19224 | 0.985203 | 7 | ATATTTGGCA | TTTGGCGC | + |
| Cebpa#MA0102.1 | 1 | 0.0181945 | 8.66056 | 0.985203 | 10 | ATATTTGGCA | GAGATTGCGAAA | - |
| SRF#MA0083.1 | 4 | 0.0194096 | 9.23895 | 0.985203 | 8 | ATATTTGGCA | GCCCATATATGG | + |
| CUP9#MA0288.1 | -1 | 0.0206445 | 9.82679 | 0.985203 | 9 | ATATTTGGCA | AATGTGTCA | - |
| MCM1#MA0331.1 | 2 | 0.0206975 | 9.85203 | 0.985203 | 10 | ATATTTGGCA | CCCAATTAGGAA | + |