MacroAPE 1083:CNhs11068 bl f2
From FANTOM5_SSTAR
Full Name: CNhs11068_bl_f2
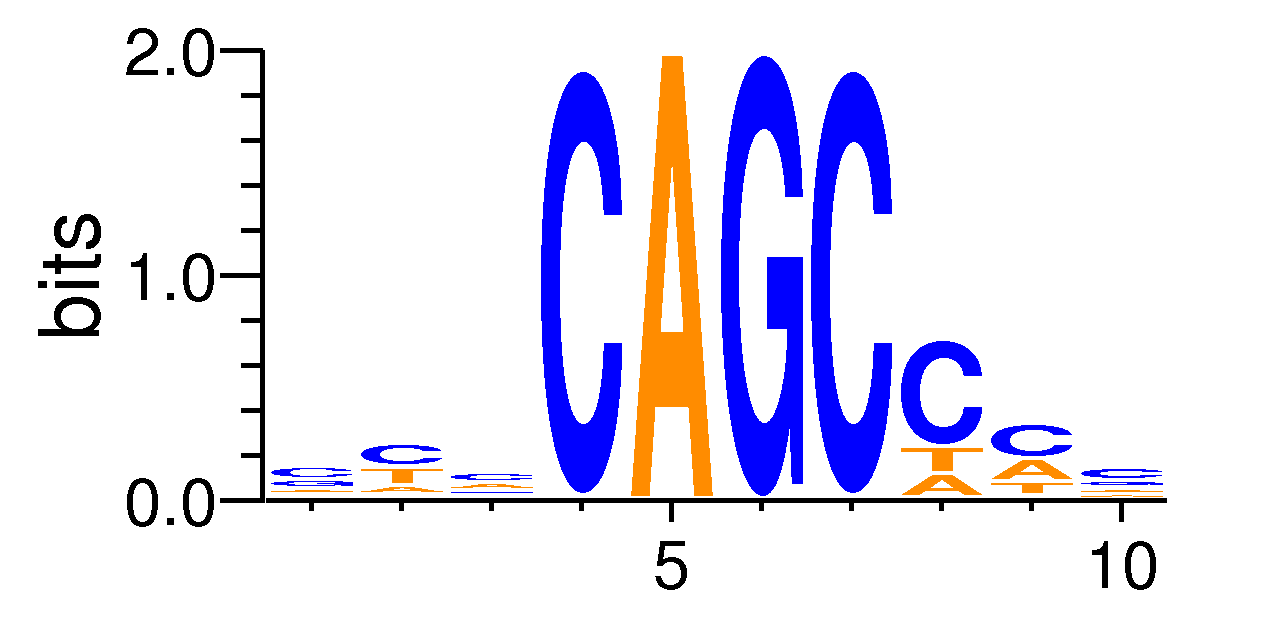
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| CRZ1#MA0285.1 | -1 | 3.47662e-05 | 0.0165487 | 0.0330541 | 9 | CCCCAGCCCC | CTAAGCCAC | + |
| SP1#MA0079.2 | 0 | 0.000694299 | 0.330486 | 0.330053 | 10 | CCCCAGCCCC | ACCCCGCCCC | - |
| ACE2#MA0267.1 | -1 | 0.00224925 | 1.07064 | 0.634776 | 7 | CCCCAGCCCC | ACCAGCA | + |
| Pax4#MA0068.1 | 15 | 0.00267062 | 1.27122 | 0.634776 | 10 | CCCCAGCCCC | GAAAAATTTCCCATACTCCACTCCCCCCCC | + |
| SWI5#MA0402.1 | 0 | 0.00395806 | 1.88404 | 0.63511 | 8 | CCCCAGCCCC | AACCAGCA | - |
| BAS1#MA0278.1 | 0 | 0.00400804 | 1.90783 | 0.63511 | 10 | CCCCAGCCCC | GCACAGCCAGAGTCAAGTCAA | + |
| SP1#MA0079.2 | -1 | 0.00718348 | 3.41934 | 0.892639 | 9 | CCCCAGCCCC | CCCCGCCCCC | + |
| Tcfcp2l1#MA0145.1 | -2 | 0.00781883 | 3.72177 | 0.892639 | 8 | CCCCAGCCCC | CCAGTTCAAACCAG | + |
| AFT2#MA0270.1 | -1 | 0.00921622 | 4.38692 | 0.892639 | 8 | CCCCAGCCCC | CACACCCC | + |
| Klf4#MA0039.2 | 1 | 0.00938875 | 4.46904 | 0.892639 | 9 | CCCCAGCCCC | GCCCCACCCA | - |
| BRCA1#MA0133.1 | -2 | 0.0155555 | 7.40441 | 0.998691 | 7 | CCCCAGCCCC | ACAACAC | + |
| btd#MA0443.1 | -2 | 0.0173602 | 8.26347 | 0.998691 | 8 | CCCCAGCCCC | TCCGCCCCCT | - |
| NHLH1#MA0048.1 | 0 | 0.0173857 | 8.27558 | 0.998691 | 10 | CCCCAGCCCC | ACGCAGCTGCGC | - |
| Zfx#MA0146.1 | 4 | 0.0210018 | 9.99687 | 0.998691 | 10 | CCCCAGCCCC | CAGGCCTCGGCCCC | - |