MacroAPE 1083:CREB1 f1
From FANTOM5_SSTAR
Full Name: CREB1_f1
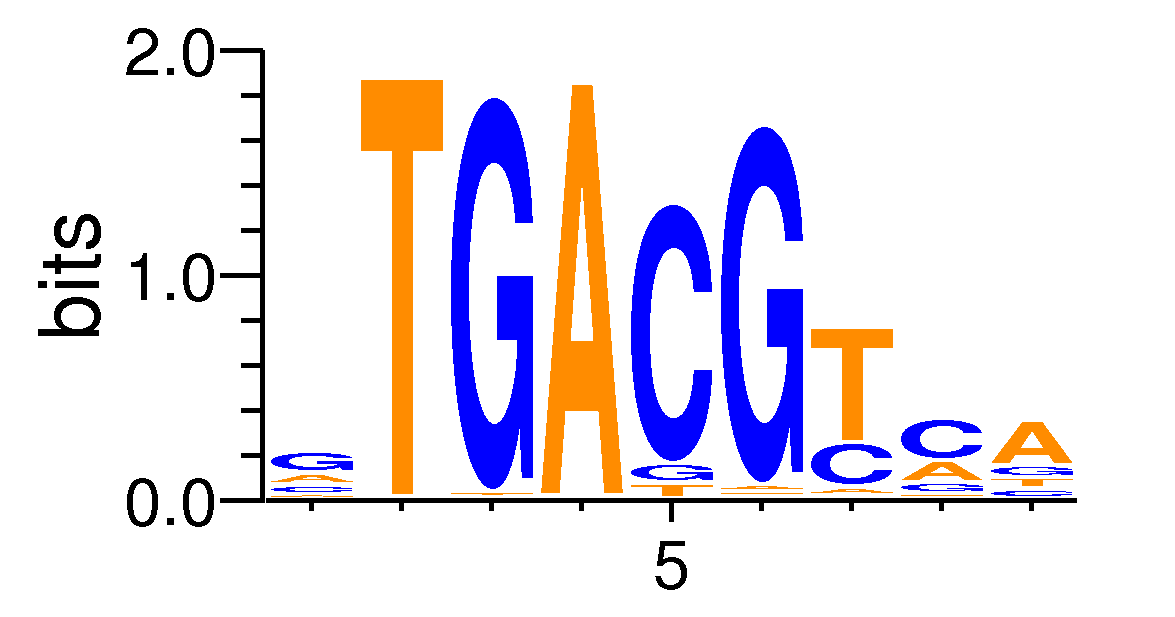
| Motif matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| CREB1#MA0018.2 | -1 | 2.021e-09 | 9.61997e-07 | 1.902e-06 | 8 | GTGACGTCA | TGACGTCA | + |
| CST6#MA0286.1 | 0 | 2.46489e-07 | 0.000117329 | 0.000115987 | 9 | GTGACGTCA | ATGACGTAA | + |
| CREB1#MA0018.2 | 4 | 1.73164e-05 | 0.00824262 | 0.0040742 | 8 | GTGACGTCA | CCTGGTGACGTC | + |
| bZIP910#MA0096.1 | 0 | 5.38609e-05 | 0.0256378 | 0.0101379 | 7 | GTGACGTCA | CTGACGT | + |
| TGA1A#MA0129.1 | -1 | 0.000169269 | 0.0805718 | 0.0265502 | 7 | GTGACGTCA | TGACGTA | - |
| HLF#MA0043.1 | 1 | 0.00130783 | 0.622527 | 0.153853 | 9 | GTGACGTCA | GGTTACGCAATT | + |
| gt#MA0447.1 | 0 | 0.00185008 | 0.88064 | 0.174115 | 9 | GTGACGTCA | ATTACGTAAT | + |
| CIN5#MA0284.1 | 1 | 0.00251931 | 1.19919 | 0.205346 | 9 | GTGACGTCA | GATTACGTAA | - |
| bZIP911#MA0097.1 | 1 | 0.00261832 | 1.24632 | 0.205346 | 9 | GTGACGTCA | GATGACGTGGCC | + |
| YAP3#MA0416.1 | 0 | 0.00843153 | 4.01341 | 0.492968 | 8 | GTGACGTCA | ATTACGTA | + |
| NR4A2#MA0160.1 | 0 | 0.0092343 | 4.39553 | 0.492968 | 8 | GTGACGTCA | GTGACCTT | - |
| Pax2#MA0067.1 | 2 | 0.00966171 | 4.59897 | 0.492968 | 6 | GTGACGTCA | GCGTGACT | - |
| Mafb#MA0117.1 | 1 | 0.0105692 | 5.03094 | 0.492968 | 7 | GTGACGTCA | GCTGACGG | + |
| YAP6#MA0418.1 | 6 | 0.0115239 | 5.48535 | 0.492968 | 9 | GTGACGTCA | GGTCTGATTACGTAAGCGAC | + |
| YPR015C#MA0435.1 | 2 | 0.0126629 | 6.02756 | 0.511988 | 9 | GTGACGTCA | TGAAGACGTAAATCCTTACA | + |
| ARG80#MA0271.1 | -1 | 0.0144181 | 6.86301 | 0.542764 | 6 | GTGACGTCA | AGACGC | + |
| NFIL3#MA0025.1 | 2 | 0.0152451 | 7.25666 | 0.542965 | 9 | GTGACGTCA | ATGTTACATAA | - |
| ARG81#MA0272.1 | 0 | 0.0155773 | 7.41479 | 0.542965 | 8 | GTGACGTCA | GTGACTCT | + |
| hth#MA0227.1 | -1 | 0.0190632 | 9.07406 | 0.630615 | 6 | GTGACGTCA | TGACAG | + |
| CUP9#MA0288.1 | -1 | 0.019432 | 9.24965 | 0.630615 | 8 | GTGACGTCA | TGACACATT | + |
| ESR2#MA0258.1 | 9 | 0.0203253 | 9.67484 | 0.637616 | 9 | GTGACGTCA | CAGGTCACCGTGACCTTG | - |