MacroAPE 1083:Motif GCTCANGG
From FANTOM5_SSTAR
Full Name: motif_GCTCANGG
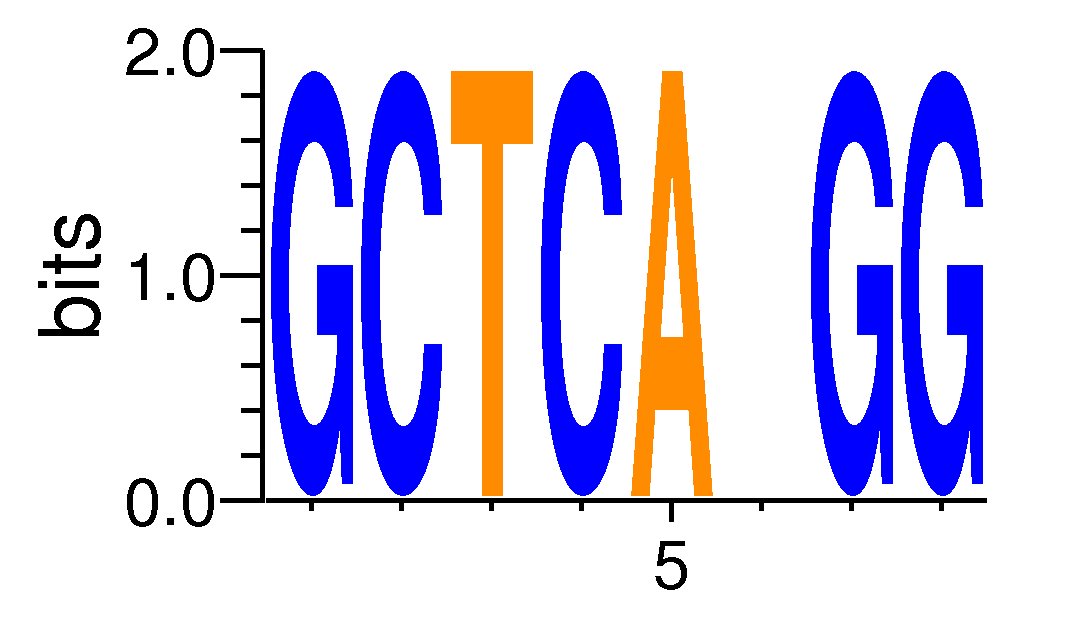
| Motif matrix | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Sample specificity
The values shown are the p-values of the motif in FANTOM5 samples. <br>Analyst: Michiel de Hoon
TomTom analysis
<br>Analyst: Michiel de Hoon and Hiroko Ohmiya
Target_ID | Optimal_offset | p-value | E-value | q-value | Overlap | Query consensus | Target consensus | Orientation |
|---|---|---|---|---|---|---|---|---|
| Esrrb#MA0141.1 | 1 | 0.00229529 | 1.09256 | 0.711783 | 8 | GCTCAAGG | AGCTCAAGGTCA | + |
| Mafb#MA0117.1 | 0 | 0.00266593 | 1.26898 | 0.711783 | 8 | GCTCAAGG | GCTGACGG | + |
| usp#MA0016.1 | 2 | 0.0032021 | 1.5242 | 0.711783 | 8 | GCTCAAGG | GGGGTCACGG | + |
| PLAG1#MA0163.1 | 3 | 0.00359427 | 1.71087 | 0.711783 | 8 | GCTCAAGG | GGGGCCCAAGGGGG | + |
| TFAP2A#MA0003.1 | 0 | 0.0037393 | 1.77991 | 0.711783 | 8 | GCTCAAGG | CCCCCGGGC | - |
| ESR2#MA0258.1 | 3 | 0.00710309 | 3.38107 | 0.775585 | 8 | GCTCAAGG | CAAGGTCACGGTGACCTG | + |
| ESR1#MA0112.2 | 2 | 0.00739245 | 3.51881 | 0.775585 | 8 | GCTCAAGG | GGGGTCACGGTGACCTGG | - |
| ESR1#MA0112.2 | 1 | 0.00741405 | 3.52909 | 0.775585 | 8 | GCTCAAGG | AGGTCAGGGTGACCTGGGCC | - |
| RXR::RAR_DR5#MA0159.1 | 1 | 0.00761694 | 3.62566 | 0.775585 | 8 | GCTCAAGG | AGGTCATGGAGAGGTCA | + |
| REI1#MA0364.1 | -2 | 0.00814897 | 3.87891 | 0.775585 | 6 | GCTCAAGG | TCAGGGG | - |
| PPARG#MA0066.1 | 3 | 0.0114832 | 5.466 | 0.96983 | 8 | GCTCAAGG | GTAGGTCACGGTGACCTACT | + |
| EBF1#MA0154.1 | 0 | 0.0124599 | 5.93093 | 0.96983 | 8 | GCTCAAGG | ACCCAAGGGA | + |
| IXR1#MA0323.1 | 4 | 0.0132468 | 6.30549 | 0.96983 | 8 | GCTCAAGG | CCCCGCTTCCGGCTT | - |
| MAC1#MA0326.1 | 3 | 0.0175237 | 8.3413 | 0.999747 | 5 | GCTCAAGG | TTTGCTCA | + |
| RXRA::VDR#MA0074.1 | 1 | 0.0179413 | 8.54005 | 0.999747 | 8 | GCTCAAGG | GGGTCAACGAGTTCA | + |
| Pax2#MA0067.1 | 0 | 0.0181157 | 8.62308 | 0.999747 | 8 | GCTCAAGG | AGTCACGG | + |